Bottleneck Removal Process in 6 Steps
with...
The program:
Variant Effect Predictor (VEP) is a Genomics program for analyzing nucleotide mutations.
VEP Stats (via Perl cloc)
- 1,431 files
- 400K lines of code
- 50 minutes for human chromosomes on a 4-core server
- compare with a 5 minutes for roughly similar Java program
Step 1. Profile using NYTProf
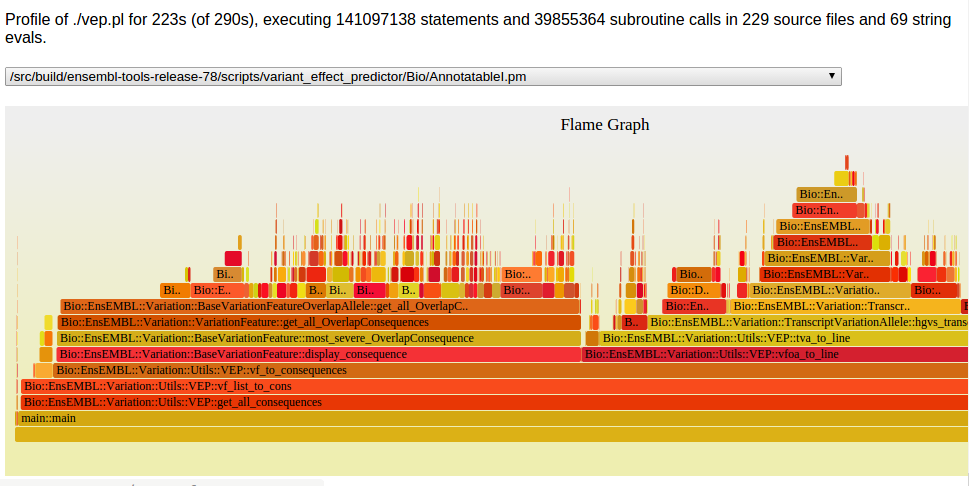
Perl Hot Spots
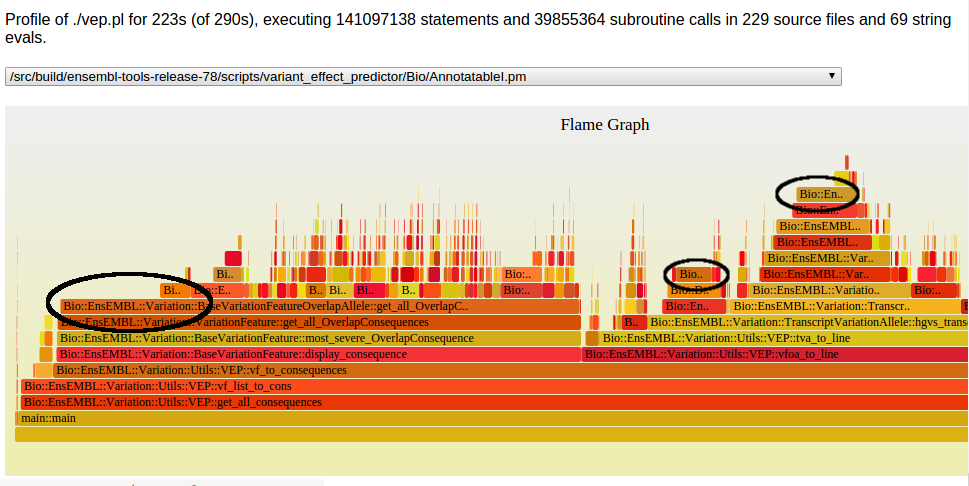 Full results are here.
Full results are here.
Slow Functions
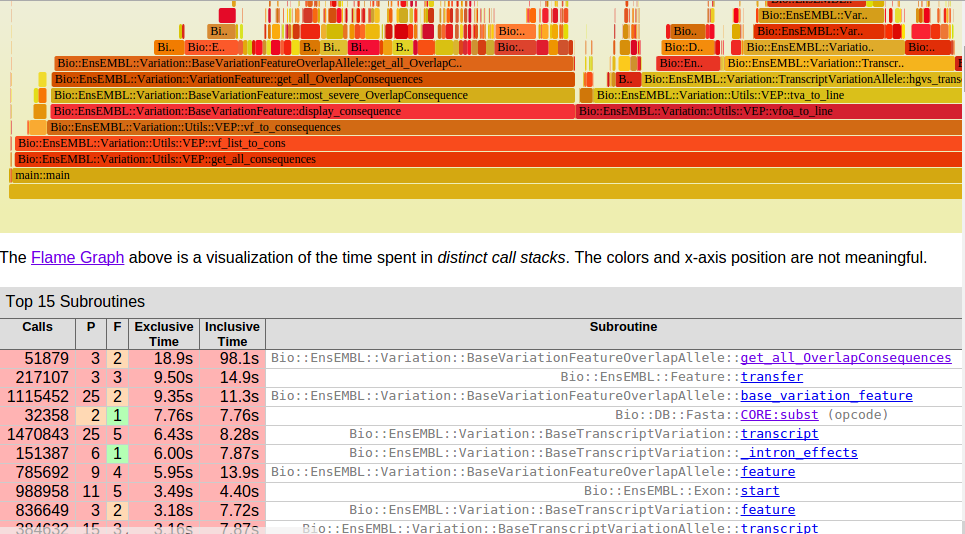
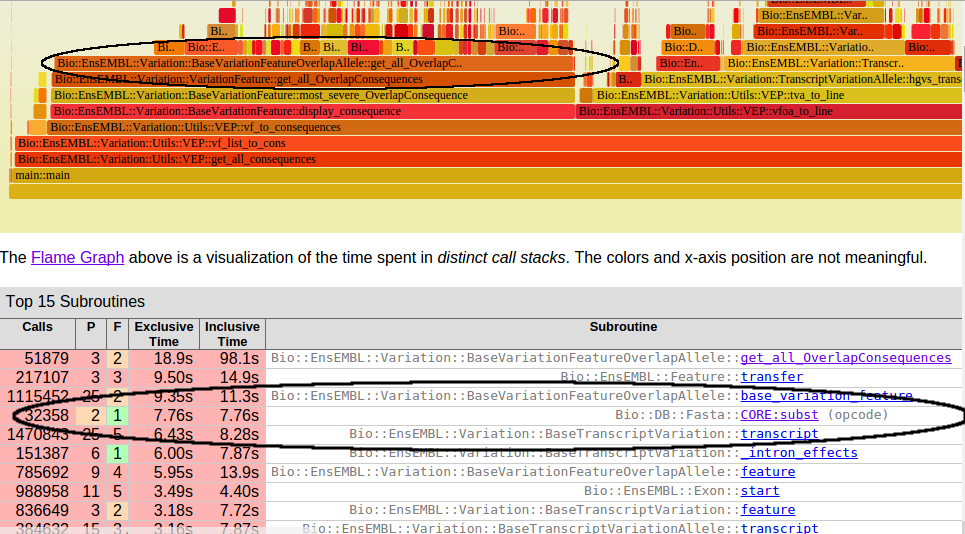
Step 2. Analyze Code
sub subseq {
...
$data =~ s/\n//g;
$data =~ s/\r//g;
...
}
Step 3. Come up with Ideas
sub subseq {
...
$data =~ s/\n//g;
}
- Compile re
- Recode in C
- ... o flag in re....
Step 4. Benchmark
use Benchmark qw(cmpthese);
cmpthese($iterations, {
'orig' => sub { ... },
're_comp' => sub { ... },
'C' => sub { ... },
});
};
A full list ideas and benchmarks is here.
Benchmark Results
perl bench.pl
Rate re_comp orig C
re_comp 800/s -- -4% -87%
orig 835/s 4% -- -86%
C 5952/s 644% 613% --
Note: results are strongly influenced by input data.
Step 5. Put in Code
my $nl = qr/\n/; my $cr = qr/\r/;
sub strip_crnl {
$_ = shift; s/$nl//g; s/$cr//g;
return $_;
}
eval q{
use Inline C => <<'END_OF_C_CODE';
char* strip_crnl(char* str) {
char *s;
char *s2 = str;
for (s = str; *s; *s++) {
if (*s != '\n' && *s != '\r') {
*s2++ = *s;
}
}
*s2 = '\0';
return str;
}
END_OF_C_CODE
};
Step 6. Run NYTProf again
- specific code:
- 7.76s => 2.23s, -5.3s (350% speedup)
- overall:
- 223s => 195s, -28s (15% speedup)
Full results for modified program are here.
Profit!
This change is now in Bio::Perl.
THE END - Thanks
- slides: https://rocky.github.io/NYC-Perl-VEP/
- rocky@gnu.org